Nov 17, 2016
Webinar: Enabling Stochastic Simulations of Movement with High Throughput Computing
Learn to run OpenSim simulations on the Open Science Grid and use probabilistic analyses to characterize your simulations
DID YOU MISS THIS EVENT?
A recording of the event is available for viewing. The example files will be made available at https://github.com/clnsmith/opensim_htc.Details
Title: Enabling Stochastic Simulations of Movement with High Throughput Computing Speakers: Colin Smith, Darryl Thelen, and Christina Koch, University of Wisconsin-Madison Time: Thursday, November 17, 2016 at 10:00 a.m. Pacific Standard TimeAbstract
Musculoskeletal simulations of movement are traditionally performed using generic models with assumed parameters and geometries. Thus, population variability and parametric uncertainty become critically important to consider when interpreting model predictions. High Throughput Computing (HTC) systems allow thousands of independent simulations to be performed in parallel, enabling probabilistic analyses to assess the sensitivities inherent in your results. We will present an overview of HTC and an HTC enabled stochastic simulation framework developed to investigate cartilage loading in the knee during movement. We will present the results of several HTC enabled studies focusing on validation in the presence of parametric uncertainty, multifactorial sensitivity analysis and structure-function relationships. Finally, we will demonstrate how to perform OpenSim simulations on the Open Science Grid, an HTC resource freely available to US researchers (and their collaborators).
Webinar highlights:
- Conceptual overview of High Throughput Computing
- Presentation of a stochastic simulation framework to investigate knee cartilage loading during movement
- Demonstration of how to perform your own OpenSim simulations on the Open Science Grid
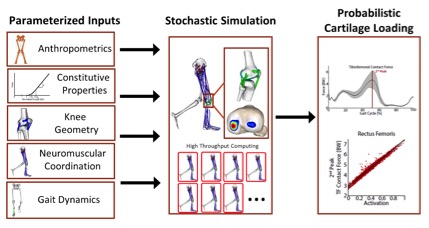
Figure 1. Overview of stochastic simulation framework to investigate knee cartilage loading during movement. By representing simulation parameters as probabilistic distributions, Smith, et al. are able to use High Throughput Computing to perform stochastic simulations to assess uncertainty propagation, parameter sensitivity and structure-function relationships.
- The University of Wisconsin-Madison Neuromuscular Biomechanics lab website
- The Center for High Throughput Computing at UW Madison
Read more about the applications discussed during the webinar:
- Lenhart, Rachel L., et al. "Prediction and validation of load-dependent behavior of the tibiofemoral and patellofemoral joints during movement." Annals of biomedical engineering 43.11 (2015): 2675-2685.
- Smith, Colin R., et al. "Influence of ligament properties on tibiofemoral mechanics in walking." Journal of Knee Surgery 29.02 (2016): 099-106.
- Smith, Colin R., et al. "The Influence of Component Alignment and Ligament Properties on Tibiofemoral Contact Forces in Total Knee Replacement." Journal of biomechanical engineering 138.2 (2016): 021017.
To start running OpenSim simulations on the Open Science Grid (OSG) yourself, you will need an account. Accounts are available for U.S. researchers, as well as international scientists who are collaborating with a U.S. scientist. Any data to be used on the Open Science Grid needs to be de-identified. They state that "Using data on the OSG is more private than having it publically available on the web, but the OSG doesn't have any specific guarantees or promises about data security." To learn more, visit the OSG website. You can also attend a User School at UW-Madison in the summer.
